Restriction Enzyme | Restriction Modification System
Table of Contents
Introduction
Restriction Enzyme or Restriction Endonuclease enzyme or Restrictase is a class of enzyme which cleaves the DNA into fragments at specific or random site on DNA molecule, known as restriction site.
like human, Bacteria is also attacked by virus (Bacteriophage : type of virus that eat bacteria). We know that virus infect by inserting its genetic material to host and to prevent it bacteria has a very interesting system called Restriction Modification System (RM).
RM System monitors the foreign DNA coming into bacterial host cell and destroy them using restriction endonuclease enzymes and at the same time protect self DNA from restriction enzyme(by modifying host DNA : methylation of DNA).
History
The Restriction Modification System origin came after studies of phage λ /bacteriophage plaque plating in early 1950. it was found that phage λ which was grown in one strain of E. coli (E. coli C), when grown in another strain (E. coli K), its yield dropped significantly. This phenomenon is called restriction, But when plating (of E.coli K) was repeated phage growth was not restricted but when inoculated to E.coli C they were once again restricted.
Based on above observation it was then said that the efficiency of phage plate depends on its particular host it was propagated, and phage that allow plating without restriction(repeating on host E.coli K) was due to modification.
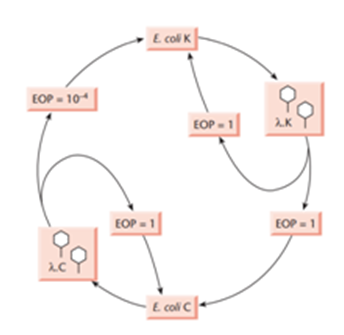
In 1962 Arber & Dussoix showed that this restriction was due to breakdown of phage DNA by host bacterial enzyme (restriction enzyme), they also showed that phage DNA insensitivity for restriction upon repeated inoculation as the result of alteration/modification of (generally methylation; Arber 1965)
In 1970 Hemilton O. Smith& Kent Wilcox discovered the restriction endonuclease enzyme (HindII) from Haemophilus influenza strain Rd. Its recognition sequence and site were elucidated by Kelly & smith on phage T7 DNA.
Leter Daniel Nathans and Kathleen Danna showed that this enzyme cleaved fragment can be separated by polyacrylamide gel electrophoresis & can be used for mapping of DNA.
In 1978 W.Arber, D. Nathans & H.O. smith awarded Noble prize of Physiology or Medicine for discovery & characterization of restriction enzyme.
Types of Restriction enzyme
Restriction enzyme (RE) categorized into four different types based their enzyme composition, cofactor requirements, and the position of restriction site relative to the recognition sequence.
- Type I RE
- Type II RE
- Type III RE
- Type IIs RE
Type I RE
- First to be identified from two different strains (K-12 and B) of E. coli.
- Type I restriction enzyme are multifunctional and are capable of both restriction digestion & modification activities and possess three subunits
- HsdR – required for restriction digestion,
- HsdM– necessary for adding methyl groups to host DNA (methyltransferase activity) and
- HsdS – for specificity of the recognition (DNA-binding) site, including restriction digestion, & methyltransferase activity
- The recognition site of Type I restriction enzyme is asymmetrical and require cofactors S-Adenosyl methionine (AdoMet), magnesium (Mg2+) ions, and energy molecule adenosine triphosphate (ATP) for activity.
- Type I restriction enzyme enzymes cleaves DNA up to 1000 bp away, from their recognition site.
Type II RE
- Type II restriction enzyme are homodimers.
- Type II restriction enzyme recognition sites are ~4–8 nucleotides in length, usually undivided and palindromic.
- Type II restriction enzyme recognize and cleave DNA at the same site at the center of both strands to yield a blunt end, or at a staggered position leaving overhangs called sticky ends.
- Type II restriction enzyme require only Mg2+ as a cofactor.
Type III RE
- Type III enzymes are hetero-oligomeric, multifunctional proteins composed of two subunits,
- Res subunits (P08764) – required for restriction digestion and
- Mod subunits (P08763) – recognises the DNA sequence specific for the system and is a modification methyltransferase.
- Type III RE cleaves DNA, ~ 20–30 base pairs away from the recognition site.
- Type III restriction enzymes require AdoMet and ATP cofactors for methylation and restriction digestion,.
- Type III enzymes recognition sites are asymmetric and leave short, single-stranded 5′ protrusions after cleavage.
Type IIs RE
- Two different enzymes but recognition sequence is asymmetric. Cleavage occurs on one side of recognition sequence up to 20 bp away
Nomenclature of Restriction enzyme
From the first identification, and characterization of restriction enzyme, till now more than 3000 restriction enzyme were identified and characterized and hundreds of them are available commercially for different purpose in the molecular lab which called for a uniform system of nomenclature.
Smith and Nathans (1973) proposed a suitable method for nomenclature of restriction enzyme- according to which (taking example of EcoRI)
- Species name of the host organism is identified by the first letter in Capital form. (Example for species Escherichia – E)
- Genus name will be represented by first two letters of the specific epithet. (Example for genus coli – first two letter co)
- A three letter (i.e. Eco) abbreviation from species and genus name is always written in italics.
- A particular strain has been identified by single letter. (e.g. strain R)
- When a particular host strain has numerous different R-M systems, these are identified by roman numerals. (I means first R-Ms system identified from E.coli)
Some examples
| Enzyme | Enzyme source |
| SmaI | Serratia marcescens, 1st enzyme |
| HindII* | Haemophilus influenzae, strain d, 2nd enzyme |
| HindIII* | Haemophilus influenzae, strain d, 3rd enzyme |
| HaeIII | Haemophilus aegyptius |
| BamHI | Bacillus amyloliquefaciens, strain H, 1st enzyme |
* here Hind (Haemophilus influenza strain d) same organism contain different R-M systems different roman number indicate their isolation sequence of that particular enzyme.
Application of Restriction enzyme
- Can be used to assist insertion of gene into vector molecule.
- Can be used to distinguish gene allele
- Can be used to digest genomic DNA for gene analysis using southern blotting
- Can be used to map genome DNA.
- Can be used to find alteration in gene.
Difference between type-I, type-II and type-III restriction enzyme
| Type-I restriction enzyme | Type-II Restriction enzyme | Type III restriction enzyme |
| hetero oligomeric | Homodimer | Hetero oligomeric |
| RE Type-I require ATP and Mg2+ for cleavage | RE Type-I require Mg2+ only for cleavage | RE Type-I require ATP for cleavage |
| In RE Type-I cleavage occur ~1000bp away from recognition site | In RE Type-III cleavage occur at or near recognition site | In RE Type-III cleavage occur away from recognition like type-I but comparatively near |
Q1. What is the restriction enzyme ?
Answer – Restiction enzyme are the special type of bacterial enzyme that cut unmodified and/or foreign DNA into fragments.
Q2. What are the examples of Restriction enzyme?
Answer – hear are the some example
EcoRI (Escherichia coli)
5′ GAATTC 3′
3′ CTTAAG 5′
BamHI (Bacillus amyloliquefaciens)
5′ GGATCG 3′
3′ CCTAGG 5′
PstI (Providencia stuartii)
5′ CTGCAG 3′
3′ GACGTC 5′
Q3. Why are restriction enzymes called molecular scissors?
Answer – Restriction enzymes are also known as molecular scissors because of it cleavage nature of DNA at molecular level at either random or specific sites.
Q.4 What is the full form of pst1 & pvu1 restriction enzymes?
Answer full form of PstI = Providencia stuartii -I and pvu = Proteus vulgaris -I.
Q5. Phage genome is not degraded by host’s endonuclease enzyme why
Answer – Because phage genome got modified when it infected the same host on second time.
1 thought on “Restriction Enzyme | Restriction Modification System”